My interests for a Ph.D. project are focused on the development and application of crop growth models, such as the APSIM model. Currently I am
learning about APSIM, DSSAT, and other crop models, applications of machine learning to these programs, and discipline-specific topics such as Whole-Genome Prediction and Genomic Selection. In the
past, I have done QTL mapping in plant populations (see publication below), and have a background in plant molecular biology and genetics.
Research Interests
- Crop Growth Models
- Genomic Prediction
- Plant Breeding
Publications
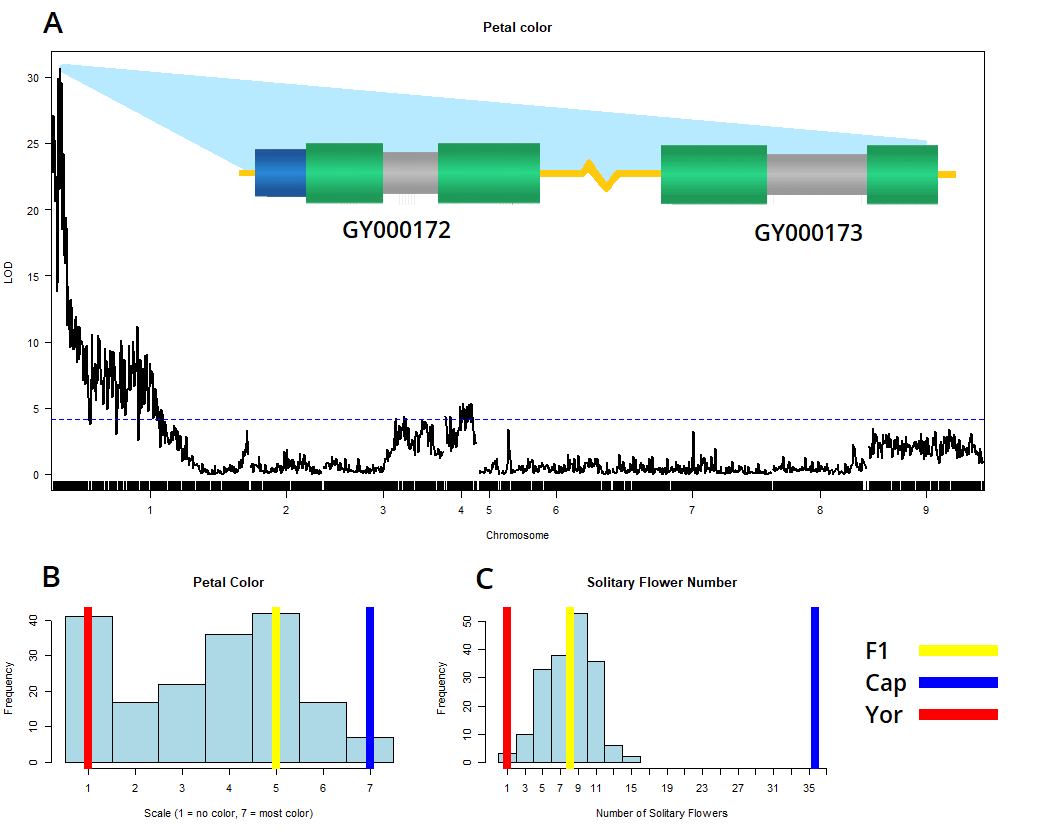
- DE Jarvis, PJ Maughan, J DeTemple, V Mosquera, Z Li, MS Barker, LA Johnson, and CJ Whipple. Chromosome-Scale Genome Assembly of Gilia yorkii Enables Genetic Mapping of Floral Traits in an Interspecies Cross Genome Biology and Evolution 14(3):evac017. March 2022. doi
Presentations
- March 2022: Utah Conference of Undergraduate Research 2022. QTL Analysis of Floral Traits in a Novel Interspecies Cross in Gilia. (Poster)
- July 2020: Botany 2020 Virtual Conference. Floral Trait Differences between Gilia yorkii and G. capitata. (Poster)
- April 2020: Library/Life Sciences Undergraduate Poster Competition 2020. Floral Traits Underlying Evolution in Gilia yorkii. (Poster)
Previous Projects
Gilia as a Model for Evo-Devo
While at BYU and working with Dr. Clint Whipple, I contributed to the development of protocols and genetic research methods for this novel model system.
This involved tissue culture, transformation via Agrobacterium and microparticle-bombardment, and EMS-mutagenesis. In addition, I conducted a QTL
analysis for floral traits on a hybrid G. yorkii x G. capitata F2 population. This model system is unique and exciting because of the
floral and inflorescence differences between the species, and the potential to discover new branching-pathway genes.
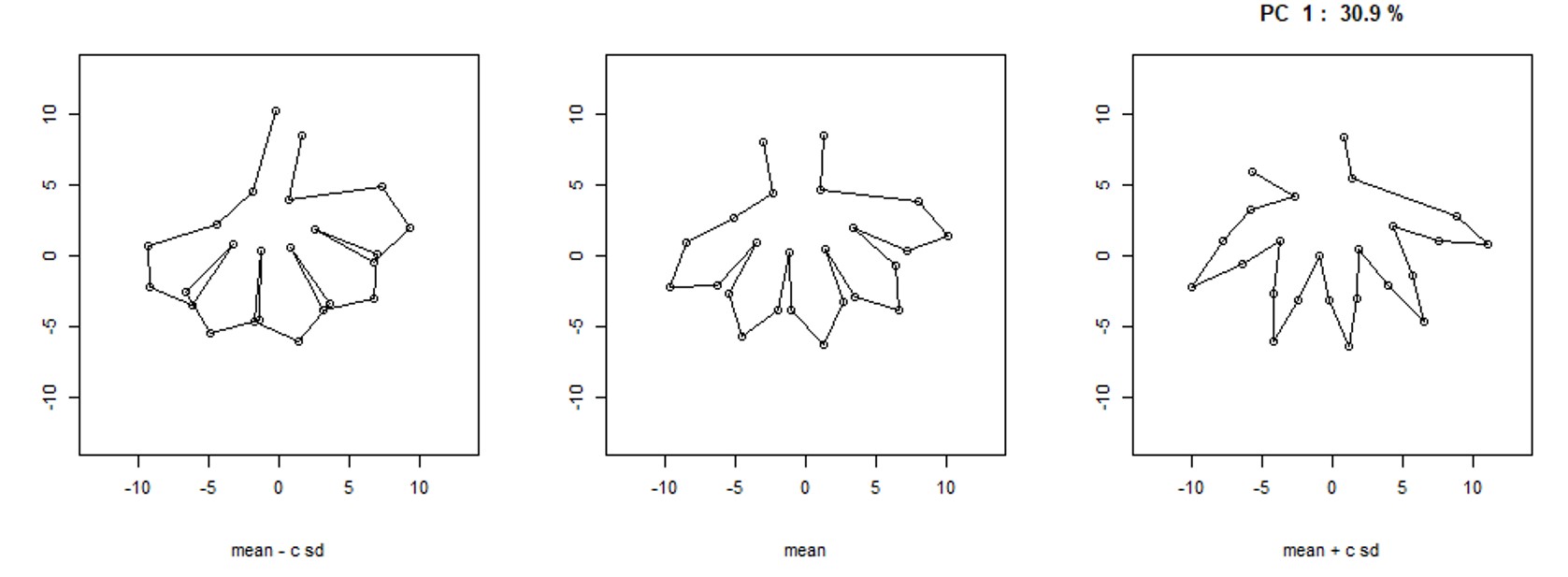
Machine-Learning approaches to Phenotyping
Under the direction of Dr. Dan Chitwood, I created training datasets from Gilia images to fit a model for Principal Component Analysis on
overall flower shape. This was an attempt to quantitatively measure overall similarity in flowers that is obvious to the human eye, but difficult
to understand through individual trait measurements. The first principal component from our model explained ~30% of the variation in the
population while capturing the observed shape differences.


